Access Superior Nucleic Acid Quantification with Digital PCR Services
Our Digital PCR expertise
As an expert dPCR service provider, our team supports any dPCR-based application, using either off-the-shelf assays or including custom primer design, all performed in an ISO 13485 accredited environment.
We developed a close collaboration with Bio-Rad and are using Bio‑Rad’s QX200™ Digital Droplet PCR™ technology.
Key applications of Digital PCR (dPCR)
dPCR enables nucleic acid quantification with superior accuracy, precision and relative sensitivity.
This technology excels in:
- Quantification of low abundant genes or small differences
- Absolute quantification
- Gene Copy Number Variation (CNV)
- Vector Copy Number (VCN) determination and Replication Competent Retrovirus (RCR) or Lentivirus (RCL) detection, in the context of adoptive cell therapies
- Rare event detection, such as cancer gene mutations in tissue or liquid biopsy
As absolute concentrations are determined, data can be easily interpreted and compared across different samples and experiments, especially in a clinical setting.
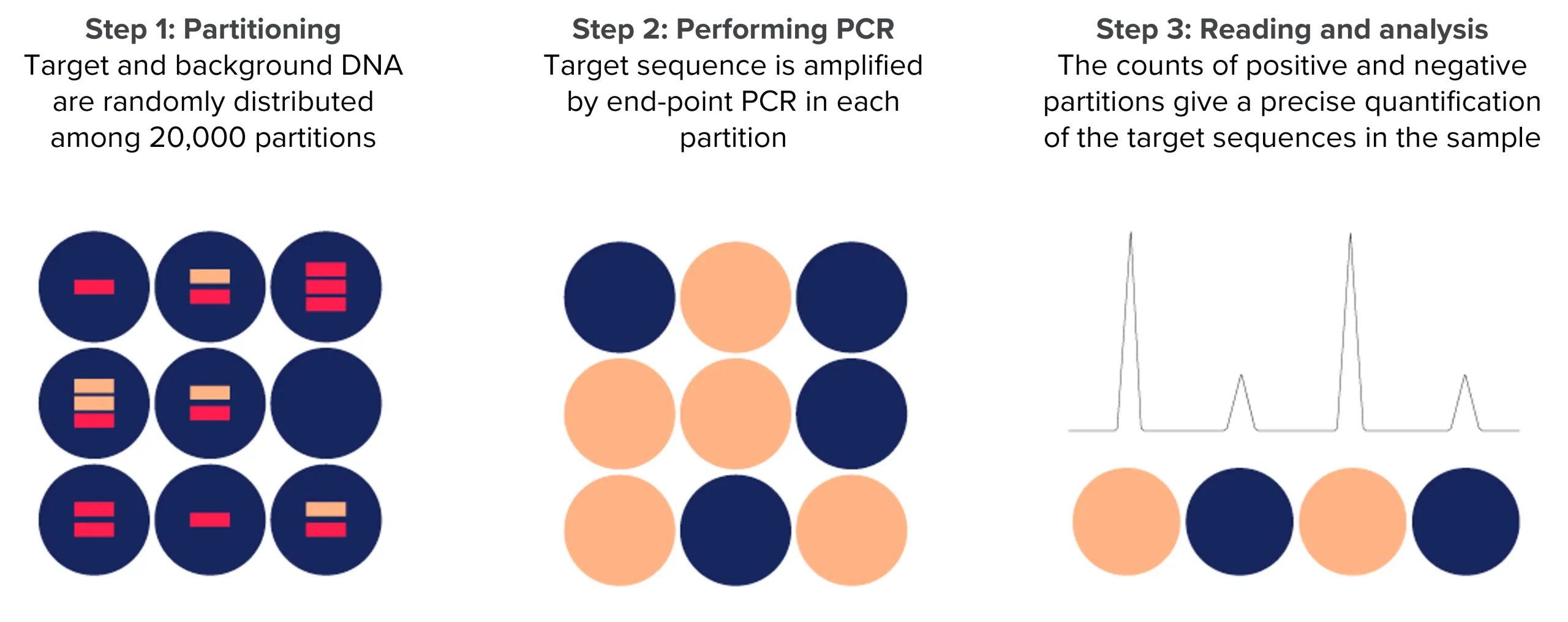
Choosing between dPCR and qPCR
Digital PCR leads to absolute quantification and does not require the use of standard curves as qPCR.
The assay particularly excels in providing the quantification of low abundant targets or small differences.
Learn more about the intrinsic potential our digital droplet PCR (ddPCR) assays in clinical studies with 4 applications performed by our team:
- Rare variant analysis in cell-free DNA (cfDNA)
- Quantification of small changes in alternative splicing
- Absolute quantification of CAR T cells biodistribution
- Absolute quantification of CAR T vectors in clinical trial
At the forefront of digital PCR based research
Our team is setting standards in the field of dPCR through participation in the development of guidelines and key scientific publications:
- The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020 (Huggett JF et al. Clin Chem, 2020)
- Quality control of digital PCR assays and platforms (Vynck M et al. Anal Bioanal Chem, 2017)
- Flexible analysis of digital PCR experiments using generalized linear mixed models (Vynck M et al. Biomol Detect Quantif, 2016)